-Search query
-Search result
Showing all 42 items for (author: ilca & sl)

EMDB-26130: 
Alpha1/BetaB Heteromeric Glycine Receptor in Strychnine-Bound State
Method: single particle / : Gibbs E, Kumar A, Chakrapani S
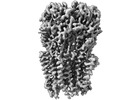
EMDB-26141: 
Alpha1/BetaB Heteromeric Glycine Receptor in Glycine-Bound State
Method: single particle / : Gibbs E, Chakrapani S, Kumar A

EMDB-11364: 
Cryo-EM structure of Spondweni virus prME
Method: single particle / : Renner M, Dejnirattisai W, Carrique L, Serna Martin I, Karia D, Ilca SL, Ho SF, Kotecha A, Keown JR, Mongkolsapaya J, Screaton GR, Grimes JM

EMDB-11366: 
Cryo-EM structure of trimeric prME spike of Spondweni virus
Method: single particle / : Renner M, Dejnirattisai W, Carrique L, Serna Martin I, Karia D, Ilca SL, Ho SF, Kotecha A, Keown JR, Mongkolsapaya J, Screaton GR, Grimes JM

EMDB-11370: 
Cryo-EM structure of mature Dengue virus 2 at 3.1 angstrom resolution
Method: single particle / : Renner M, Dejnirattisai W, Carrique L, Serna Martin I, Karia D, Ilca SL, Ho SF, Kotecha A, Keown JR, Mongkolsapaya J, Screaton GR, Grimes JM

EMDB-11371: 
Cryo-EM structure of mature Spondweni virus
Method: single particle / : Renner M, Dejnirattisai W, Carrique L, Serna Martin I, Karia D, Ilca SL, Ho SF, Kotecha A, Keown JR, Mongkolsapaya J, Screaton GR, Grimes JM

EMDB-11372: 
Cryo-EM structure of immature Spondweni virus
Method: single particle / : Renner M, Dejnirattisai W, Carrique L, Serna Martin I, Karia D, Ilca SL, Ho SF, Kotecha A, Keown JR, Mongkolsapaya J, Screaton GR, Grimes JM

EMDB-11008: 
Icosahedral reconstruction of human adenovirus D26 capsid
Method: single particle / : Huiskonen JT, Abrishami V

EMDB-11012: 
Localized reconstruction of human adenovirus D26 hexon type 3
Method: single particle / : Huiskonen JT, Abrishami V

EMDB-11009: 
Localized reconstruction of the trimeric fibre and its interactions with the penton base of human adenovirus HAdV-D26
Method: single particle / : Huiskonen JT, Abrishami V

EMDB-11010: 
Localized reconstruction of human adenovirus D26 hexon type 1
Method: single particle / : Huiskonen JT, Abrishami V

EMDB-11011: 
Localized reconstruction of human adenovirus D26 hexon type 2
Method: single particle / : Huiskonen JT, Abrishami V

EMDB-11013: 
Localized reconstruction of human adenovirus D26 hexon type 4
Method: single particle / : Huiskonen JT, Abrishami V

EMDB-10289: 
Bacteriophage phi6 dsRNA genome, layer 1, conformation pseudo C2
Method: single particle / : Ilca SL, Huiskonen JT

EMDB-10075: 
Bacteriophage phi6 dsRNA genome, layer 1, conformation pseudo D3
Method: single particle / : Ilca SL, Huiskonen JT

EMDB-0294: 
Bacteriophage phi6 dsRNA genome, layer 2
Method: single particle / : Ilca SL, Huiskonen JT

EMDB-0295: 
Bacteriophage phi6 dsRNA genome, layer 3
Method: single particle / : Ilca SL, Huiskonen JT

EMDB-0296: 
Bacteriophage phi6 dsRNA genome, layers 4 and 5
Method: single particle / : Ilca SL, Huiskonen JT

EMDB-0299: 
Bacteriophage phi6 nucleocapsid reconstructed with icosahedral symmetry
Method: single particle / : Ilca SL, Huiskonen JT

EMDB-0300: 
Reconstruction of dsRNA bacteriophage phi6 nucleocapsid with D3 symmetry
Method: single particle / : Ilca SL, Huiskonen JT

EMDB-0302: 
Bacteriophage phi6 dsRNA genome, layer 1, conformation pseudo D3'
Method: single particle / : Ilca SL, Huiskonen JT

EMDB-0304: 
Bacteriophage phi6 dsRNA genome, layer 1, conformation pseudo D3', sub-conformation 1
Method: single particle / : Ilca SL, Huiskonen JT

EMDB-0305: 
Bacteriophage phi6 dsRNA genome, layer 1, conformation pseudo D3', sub-conformation 2
Method: single particle / : Ilca SL, Huiskonen JT

EMDB-0306: 
Bacteriophage phi6 dsRNA genome, layer 1, conformation pseudo D3', sub-conformation 3
Method: single particle / : Ilca SL, Huiskonen JT

EMDB-4633: 
Cryo-EM structure of SH1 full particle.
Method: single particle / : De Colibus L, Roine E, Walter TS, Ilca SL, Wang X, Wang N, Roseman AM, Bamford D, Huiskonen JT, Stuart DI

EMDB-4634: 
Localized reconstruction of archaeal virus SH1 vertex calculated without symmetry
Method: single particle / : De Colibus L, Ilca SL, Stuart DIS, Huiskonen JT

EMDB-4656: 
Localized reconstruction of archaeal virus SH1 spike calculated with C2 symmetry
Method: single particle / : De Colibus L, Ilca SL, Stuart DIS, Huiskonen JT

EMDB-0245: 
Structure of the in vitro assembled bacteriophage phi6 P1P4 complex
Method: single particle / : Huiskonen JT, Ilca SL

EMDB-3630: 
CryoEM Structure of Foot and Mouth Disease Virus O PanAsia
Method: single particle / : Kotecha A, Stuart D

EMDB-3631: 
CryoEM Structure of Foot and Mouth Disease Virus O1 Manisa
Method: single particle / : Kotecha A, Stuart D

EMDB-3632: 
Localised reconstruction of alpha v beta 6 bound to Foot and Mouth Disease Virus O PanAsia - Pose A.
Method: single particle / : Kotecha A, Stuart D

EMDB-3633: 
Localised reconstruction of alpha v beta 6 bound to Foot and Mouth Disease Virus O PanAsia - Pose A prime.
Method: single particle / : Kotecha A, Stuart D

EMDB-3634: 
Localised Reconstruction of Integrin alpha V beta 6 bound to Foot and Mouth Disease Virus O1 Manisa - Pose A.
Method: single particle / : Kotecha A, Stuart D

EMDB-3635: 
Localised Reconstruction of Integrin alpha V beta 6 bound to Foot and Mouth Disease Virus O1 Manisa - Pose B.
Method: single particle / : Kotecha A, Stuart D

EMDB-3571: 
dsRNA bacteriophage phi6 nucleocapsid
Method: single particle / : Sun Z, El Omari K, Sun X, Ilca SL, Kotecha A, Stuart DI, Poranen MM, Huiskonen JT

EMDB-3572: 
Localized reconstruction of bacteriophage phi6 packaging hexamer P4
Method: single particle / : Sun Z, El Omari K, Sun X, Ilca S, Kotecha A, Stuart DI, Poranen MM, Huiskonen JT

EMDB-3573: 
Localized reconstruction of bacteriophage phi6 vertex
Method: single particle / : Sun Z, El Omari K, Sun X, Ilca S, Kotecha A, Stuart DI, Poranen MM, Huiskonen JT
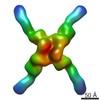
EMDB-3183: 
Localized reconstruction of Sec13/31 vertex from dodecahedral COPII cage
Method: single particle / : Ilca S, Kotecha A, Sun X, Poranen MP, Stuart DI, Huiskonen JT

EMDB-3184: 
Localized reconstruction of rotavirus VP4 spike
Method: single particle / : Ilca S, Kotecha A, Sun X, Poranen MP, Stuart DI, Huiskonen JT

EMDB-3185: 
Structure of the in vitro assembled bacteriophage phi6 polymerase complex
Method: single particle / : Ilca S, Kotecha A, Sun X, Poranen MP, Stuart DI, Huiskonen JT

EMDB-3186: 
Structure of the P2 polymerase inside in vitro assembled bacteriophage phi6 polymerase complex
Method: single particle / : Ilca S, Kotecha A, Sun X, Poranen MP, Stuart DI, Huiskonen JT

EMDB-3187: 
Structure of the P2 polymerase inside in vitro assembled bacteriophage phi6 polymerase complex, with P1 included
Method: single particle / : Ilca S, Kotecha A, Sun X, Poranen MP, Stuart DI, Huiskonen JT
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
